- >>
- Healthy Adult Studies
- HCP Young Adult
- Project Protocols Detail
Components of the Human Connectome Project - Informatics
Informatics is an increasingly important enabler of the scientific process. The overarching mission of the HCP informatics team is to develop, deploy, and share an informatics platform that maximizes the scientific community’s ability to utilize HCP data.
This platform includes tools to support the HCP across the totality of the project, from data acquisition to data sharing. The HCP platform includes two interoperable components: ConnectomeDB, a data management system, and Connectome Workbench, a suite of scientific productivity tools. ConnectomeDB is based on XNAT, a widely used imaging informatics platform developed by the Neuroinformatics Research Group. ConnectomeDB is used internally by HCP operations groups to capture and process data and to perform quality control procedures. It will also be used as the primary mechanism for data dissemination. Connectome Workbench is based on Caret, a desktop application for brain mapping. Workbench will provide users with visualization and discovery tools that interact seamlessly with ConnectomeDB.
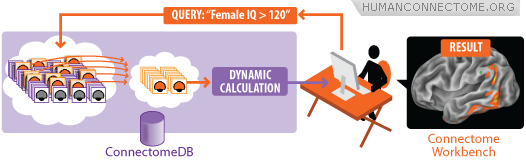
In addition to developing software, the informatics team leads several other components of the HCP. It oversees a high performance computing resource on which the HCP’s computational processes are executed. It oversees the overall workflow of data acquisition, ensuring that a global quality assurance process is maintained and appropriate privacy and security safeguards are in place. It works closely with external informatics resources, such as the Allen Brain Institute and the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC), to ensure that HCP data are integrated with complementary databases and analytic tools. And it guides the development of data sharing policies, procedures, and documentation to enable rapid and effective dissemination of HCP data.
HCP Informatics operates on 3 basic principles:
- Usability: We follow user centered software engineering practices to build software that accommodates the full range of HCP users. The informatics team includes a full time user-experience engineer and is guided by a user-experience committee that includes both end users and user-experience experts.
- Interoperability: All software is built to interoperate with the emerging biomedical informatics backbone being developed by the BIRN, caBIG, and other informatics organizations. We also conform to emerging neuroimaging data standards (NIFTI, GIFTI) as well as the DICOM standard for initial image acquisition. Team members have a strong track record in building interoperable software and in contributing to this informatics backbone. This approach ensures that users across the research spectrum have full access to the HCP data.
- Openness: All data, protocols, documentation, and other materials will be shared according to an open access standard defined by Open Knowledge Foundation (OKF). Source code for the informatics platform and associated software will be released under the open source standard defined by the Open Source Initiative (OSI).