Using Connectome Workbench
Connectome Workbench is an open source, freely available visualization and discovery tool used to map neuroimaging data, especially data generated by the Human Connectome Project. The distribution includes wb_view, a GUI-based visualization platform, and wb_command, a command-line program for performing a variety of algorithmic tasks using volume, surface, and grayordinate data. wb_command is necessary for running HCP data processing pipelines.
Workbench version 2.1.0 is available for download for 64-bit Mac OSX, Windows, and Linux: Get Connectome Workbench now.
Changes in Workbench v2.1.0
Changes in Workbench 2.1.0 are listed on Github: View changes
This release fixes a bug in upload of scenes to the BALSA database.
About Connectome Workbench
Workbench allows you to explore data and activity on the surface, as well as in the volume of the brain. wb_view works seamlessly with ConnectomeDB to import averaged data from a group of subjects and view that data on a group averaged brain atlas in Workbench.
wb_command allows you to perform a variety of algorithmic tasks using volume, surface, and grayordinate data. Information on the use of the included commands is on the wb_command documentation page.
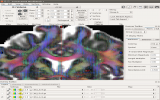
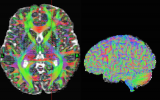
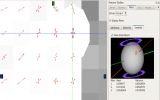
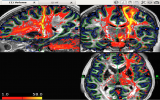
Screenshots (click to expand)
Software Platform
The Workbench visualization platform and wb_command utilities are open-source and written in C++ by software developers in the Van Essen Laboratory at Washington University. (License)
Release Dates
Connectome Workbench v2.1.0 update has been released as of June 5, 2025 and is available for MacOSX, Linux, Windows platforms.
Related Data and Documentation Releases:
- Workbench 1.5 Tutorial (PDF)
Download Workbench 1.0-1.4.2 Tutorial - Guide to Workbench Annotations (PDF)
- WB tutorial & user guide and HCP sample data available from BALSA. Register for a BALSA account and agree to Data Use Terms to download.
- WB-compatible HCP-YA Group average functional connectivity dataset and dense connectome files are available from ConnectomeDB
Source Code
Download the source code for Connectome Workbench from our GitHub repository: https://github.com/Washington-University/workbench.