- >>
- Healthy Adult Studies
- HCP Young Adult
- News
- Article
Announcing the HCP 900 Subjects Data Release
 The Human Connectome Project (HCP) WU-Minn consortium is pleased to announce the release of 900 Subjects of HCP image and behavioral data, including 95 MEG datasets.
The Human Connectome Project (HCP) WU-Minn consortium is pleased to announce the release of 900 Subjects of HCP image and behavioral data, including 95 MEG datasets.
What’s in the HCP 900 Subjects data release? The 900 Subjects release includes behavioral/demographic and 3T MR imaging data from 970 healthy adult participants collected in 2012-spring 2015. Structural scans are available for 897 subjects. 730 subjects completed all of the four MRI modalities in the HCP protocol: structural images (T1w and T2w), resting-state fMRI (rfMRI), task fMRI (tfMRI), and high angular resolution diffusion imaging (dMRI). In addition to MR scans, 95 subjects also have at least some resting-state MEG (rMEG) and/or task MEG (tMEG) data available, with 50 subjects completing the entire HCP MEG protocol.
The 900 Subjects release also includes:
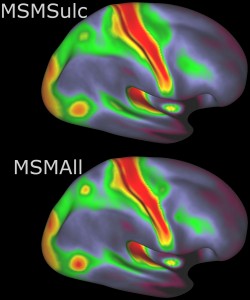
Addition of MSM-All registration and data in minimally preprocessed and analysis packages. In addition to MSM-Sulc (cortical folding only-based registration) introduced in the S500 release, the structural MRI processing pipeline now also performs intersubject registration using individual cortical folding, myelin map, and rfMRI correlation data together (MSM-All) to a group-average surface template via a multimodal surface matching (MSM) algorithm (Robinson et al. 2014, Smith et al. 2013).
ConnectomeDB offers the “MSM-Sulc + MSM-All” package type for all S900 subjects and the “S500 to S900 Extension Only” packages to “patch” users’ existing S500 data with the MSM-All and other added files available for the S900 release.
Resting State fMRI FIX-Denoised (Extended) packages expanded. FIX-Denoised (Extended) rfMRI packages have been expanded to contain a “stats.dscalar.nii” and a “RestingStateStats” folder. These files provide information about different types of ‘noise’ and ‘signal’ in HCP resting state data, gleaned by partitioning the variance according to different processing stages in the FIX denoising pipeline.
Physiological data files corrected. We have corrected timing errors in the processed physiological log files for each scan that resulted in variable offsets in timing. These corrected .txt files are in the S900 data packages.
Source-level rMEG and tMEG data expanded to include beamformer connectivity results. New source-level processing pipelines using the Beamformer filters inverse algorithms have been implemented to estimate time-resolved dense connectomes in a number of frequency bands.
Parcellated rMEG band limited power envelope and connectivity results now available. Results of the Icablpenv, Icablpcorr, icaimagcoh, and bfblpenv pipelines are additionally available in parcellated versions that were created using the Yeo et al. 2011 17 network parcellation.
Soon to be available:
- Updated group-average rfMRI dense connectivity and tfMRI data. Group-average rfMRI dense connectivity data and group-average analyzed task data for an 800+ group of S900 subjects with complete rfMRI data and tfMRI data is currently being prepared for release in December 2015. These data will be released as Connectome Workbench-compatible datasets.
- Updated parcellation, timecourse, and netmap (PTN) data. Individual PTN data for all S900 subjects with complete rfMRI data (800+ subjects) is also being prepared for release.
All S900 imaging data soon to be available on the cloud through Amazon S3. HCP has continued our partnership with the Amazon Web Services Amazon Web Services (AWS) Public Data Sets program (http://aws.amazon.com/publicdatasets/) to offer storage and access to all HCP S900 imaging data on Amazon S3 within weeks of the 900 Subjects Release. (Currently, S500 data is still available via S3)
Access 900 Subjects data on the HCP website. Explore, download, or order the HCP 900 Subjects dataset (~17TB of data!) via the ConnectomeDB database. Most HCP image and behavioral data is openly accessible to investigators worldwide who register and accept a limited set of Open Access Data Use Terms. Note: Please clear your browser cache before logging in to ConnectomeDB.
Want more information? Check out the HCP 900 Subjects Release Reference Manual for a comprehensive guide that includes details on imaging protocols, behavioral measures, and information that will help users obtain and analyze the 900 Subjects data.
If you are actively using HCP data and tools, we encourage you to join and be active in the hcp-users discussion group (http://www.humanconnectome.org/contact/#subscribe), so that you can tune in to technical discussions on issues that may be of interest.