- >>
- Healthy Adult Studies
- HCP Young Adult
- News
- Article
HCP Announces Release of Data from 500 Subjects
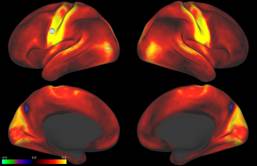
The Human Connectome Project (HCP) WU-Minn consortium is pleased to announce the release of 500 Subjects of HCP image and behavioral data.
What’s in the HCP 500 Subjects data release? The 500 Subjects release includes behavioral/demographic and 3T MR imaging data from 523 healthy adult participants collected in Q1-Q6. Structural scans are available for over 500 subjects. Over 440 subjects completed all of the four imaging modalities in the HCP protocol: structural images (T1w and T2w), resting-state fMRI (rfMRI), task fMRI (tfMRI), and high angular resolution diffusion imaging (dMRI). In addition to MR scans, 14 subjects were also scanned in resting-state MEG (rMEG) and task MEG (tMEG).
The 500 Subjects release also includes:
Significant updates for MR minimal preprocessing pipelines. Changes have been made to all of the HCP minimal preprocessing pipelines, including a switch to MSMSulc registration from FreeSurfer registration in structural MR pipeline, component analysis software updates (FSL 5.0.6, FreeSurfer 5.3.0-HCP, Connectome Workbench beta v0.85), output files in CIFTI-2.0 format, . An update to the CIFTI file format was released in March 2014 (see https://www.nitrc.org/projects/cifti/ for details). All CIFTI files produced by the HCP pipelines are now in CIFTI-2.0 format.
Individual task fMRI analysis results available at various surface smoothing levels. Individual (within-subject) grayordinates (surface)-based cross run level-2 tfMRI analysis results using FSL are now available at several levels of total smoothing: 2mm, 4mm, 8mm, and 12mm smoothed on the surface. 4mm volume-smoothed cross run tfMRI analysis results are also available.
Updates to MEG data and access in ConnectomeDB. Cortical sheet based anatomical source models are now available, MEG data have been integrated into the ConnectomeDB dashboard, and MR data is now available for all released MEG subjects.
Group-average functional MR data for two larger subject groups. Resting state functional connectivity MRI (fcMRI) correlation matrices (“dense” functional connectome files), in full correlation matrix and mean gray signal regressed versions are available for a group of 468 ‘related’ subjects (R468) with complete rfMRI data. Task functional MRI (tfMRI) contrasts are available using data processed with the updated processing pipelines for two groups: 100 unrelated (U100) subjects and 400+ ‘related’ subjects (R400+) that are a mixture of related and unrelated subjects.
Additional restricted behavioral and demographic data. Restricted data has been added for all subjects from alcohol and tobacco 7-day retrospective surveys; demographic, mental health, and substance use data from the semi-structured assessment for the genetics of alcoholism (SSAGA) interview; physical health and history measures; menstrual cycle information on female subjects; and family history of psychiatric illness.
Improvements to behavioral data organization and data dictionary. We have reorganized fields, renamed variables, and improved descriptions for behavioral and individual difference data to make navigation and filtering more intuitive in ConnectomeDB.
All imaging data soon to be available on the cloud through Amazon S3. HCP has partnered with Amazon Web Services Amazon Web Services (AWS) Public Data Sets program (http://aws.amazon.com/publicdatasets/) to offer storage and access to all HCP imaging data on Amazon S3 within weeks of the 500 Subjects Release.
Access 500 Subjects data on the HCP website. Explore, download, or order the HCP 500 Subjects dataset (~17TB of data!) via the ConnectomeDB database. Most HCP image and behavioral data is openly accessible to investigators worldwide who register and accept a limited set of Open Access Data Use Terms. Note: Please clear your browser cache before logging in to ConnectomeDB.
Want more information? Check out the HCP 500 Subjects Release Reference Manual for a comprehensive guide that includes details on imaging protocols, behavioral measures, and information that will help users obtain and analyze the 500 Subjects data.
If you are actively using HCP data and tools, we encourage you to join and be active in the hcp-users discussion group (http://www.humanconnectome.org/contact/#subscribe), so that you can tune in to technical discussions on issues that may be of interest.
Thanks again for your interest in the HCP. Please send us your questions and comments anytime to info@humanconnectome.org.